Using PyRigi for Rigidity Theory¶
This notebook illustrates how to use PyRigi for applications in rigidity theory using the
classes Graph and Framework.
It can be downloaded here.
import pyrigi.frameworkDB as frameworks
import pyrigi.graphDB as graphs
from pyrigi import Graph, Framework
Framework Construction¶
One way to construct a framework is to provide a graph and a
realization to the constructor
Framework. For instance, a framework F1 on the complete graph on 4 vertices \(K_4\)
can be constructed in the following way.
K4 = graphs.Complete(4)
F1 = Framework(K4, {0:[1,2], 1:[0,5], 2:[-1,'1/2 * sqrt(5)'], 3:['1/2','4/3']})
F1
Framework(Graph.from_vertices_and_edges([0, 1, 2, 3], [(0, 1), (0, 2), (0, 3), (1, 2), (1, 3), (2, 3)]), {0: ['1', '2'], 1: ['0', '5'], 2: ['-1', 'sqrt(5)/2'], 3: ['1/2', '4/3']})
It is particularly important to provide the numbers as sympy expressions or strings if you intend to
perform symbolic computations. Otherwise, fractions are converted to floating point numbers, which may
cause issues with the symbolic computation of rigidity properties for the framework. The following
example illustrates this behavior.
Framework(graphs.Complete(1), {0:[1/2,2/3]})
Framework(Graph.from_vertices_and_edges([0], []), {0: ['0.500000000000000', '0.666666666666667']})
If you want to use floating point coordinates and you do not specify that the computation should be numeric
via the keyword numerical=True in the appropriate methods, then incorrect results may occur.
A framework can then be visualized by calling the method plot() on F1,
see also tutorial Plotting.
F1.plot()
The position of a vertex in F1 can be retrieved by calling
F1[2]
Class methods¶
Alternatively, the realization can be specified by using a class method. For instance, Simplicial() creates
a realization on the \(d\)-simplex.
F2 = Framework.Simplicial(K4, 3)
F2
Framework(Graph.from_vertices_and_edges([0, 1, 2, 3], [(0, 1), (0, 2), (0, 3), (1, 2), (1, 3), (2, 3)]), {0: ['0', '0', '0'], 1: ['1', '0', '0'], 2: ['0', '1', '0'], 3: ['0', '0', '1']})
The dimension that a framework is embedded in can be accessed via the dim() method.
F2.dim
3
F2[2]
The framework can be translated using the translate() method.
F2.translate([3,4,5])
F2[2]
Framework database¶
There exists a framework database from which certain frameworks can be imported. A detailed tutorial of it can be accessed here.
Rigidity properties¶
Infinitesimal rigidity¶
One of the main applications of PyRigi is to determine whether a framework is
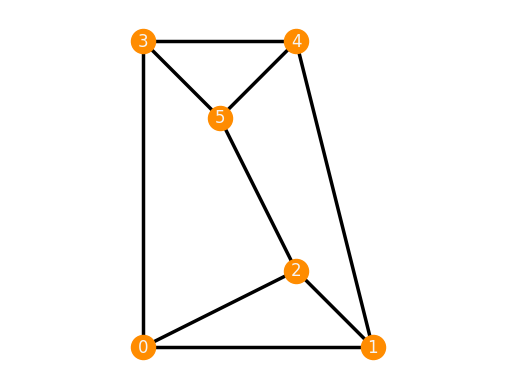
infinitesimally rigid. Take for example the following realization of the 3-prism.
We can determine whether it is infinitesimally rigid using the method is_inf_rigid().
TP_rigid = frameworks.ThreePrism()
TP_rigid.plot()
TP_rigid.is_inf_rigid()
True
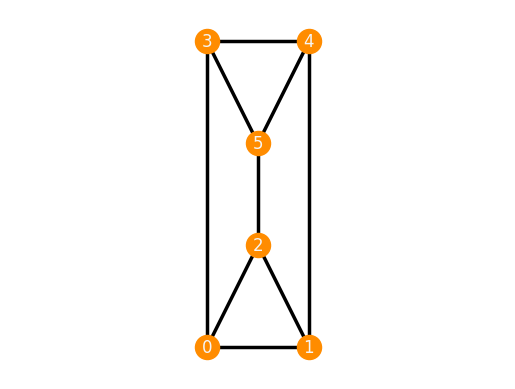
We can also create an infinitesimally flexible, but continuously rigid realization of the 3-prism using the parameter 'parallel'.
TP_parallel = frameworks.ThreePrism('parallel')
TP_parallel.plot()
TP_parallel.is_inf_rigid()
False
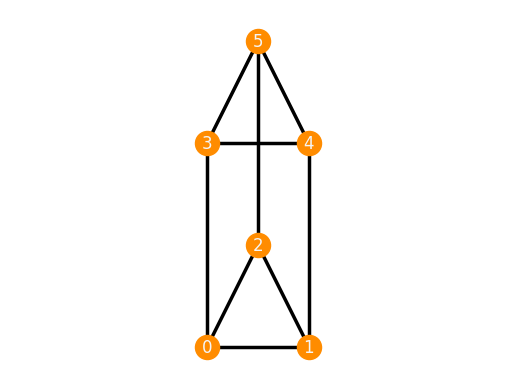
We check its rigidity using the method is_inf_rigid().
Finally, a continuously flexible realization can be created using the keyword 'flexible'.
TP_flexible = frameworks.ThreePrism('flexible')
TP_flexible.plot()
TP_flexible.is_inf_rigid()
False
Adding an edge to the 3-prism changes its rigidity properties.
TP_e = frameworks.ThreePrismPlusEdge()
TP_e.plot()
TP_e.is_inf_rigid()
True
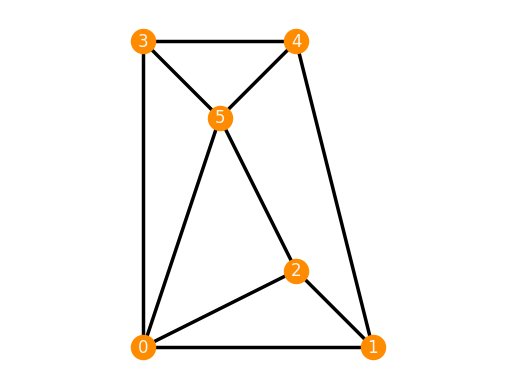
In particular, the resulting framework is not minimally infinitesimally rigid anymore, even though the 3-prism was.
print(TP_rigid.is_min_inf_rigid())
print(TP_e.is_min_inf_rigid())
True
False
To investigate further properties of the framework, PyRigi can be used for computing the corresponding
rigidity matrix using the method rigidity_matrix().
TP_flexible.rigidity_matrix()
If you do not want to compute the infinitesimal flexes on your own, you can ask PyRigi to do it. The method
nontrivial_inf_flexes() returns a list of nontrivial infinitesimal flexes
that all in the format of a matrix.
inf_flexes = TP_flexible.nontrivial_inf_flexes()
inf_flexes[0]
We can verify that a vector is indeed an nontrivial infinitesimal flexes by calling the method
is_nontrivial_flex().
print(TP_flexible.is_nontrivial_flex(inf_flexes[0]))
print(TP_rigid.is_nontrivial_flex(inf_flexes[0]))
True
False
The list of trivial infinitesimal flexes can be accessed via the method
trivial_inf_flexes().
inf_flexes = TP_flexible.trivial_inf_flexes()
inf_flexes[0]
Equilibrium Stresses¶
PyRigi can also be used to compute equilibrium stresses.
This is done via the method stresses().
F = frameworks.Frustum(3)
inf_flex = F.inf_flexes()[0]
stress = F.stresses()[0]
stress
We can visualize both the infinitesimal flexes and equilibrium stresses of a framework by using the appropriate keywords.
F.plot(inf_flex=inf_flex, stress=stress)
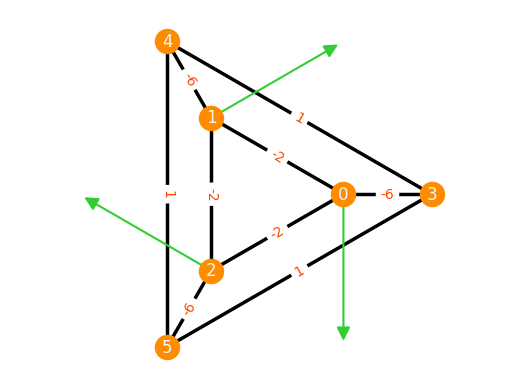
Again, it can be verified that the stress indeed lies in the cokernel of the rigidity matrix by calling
is_stress().
F.is_stress(stress)
True
The stress matrix associated to a framework for a given stress can be accessed via
the method stress_matrix().
Omega = F.stress_matrix(stress)
Generic rigidity¶
We can also use PyRigi to investigate the generic rigidity of graphs.
The underlying graph of a framework can be accessed
via the property Framework.graph and the rigidity of this graph can be determined via
is_rigid().
G_TP = TP_rigid.graph
G_TP.is_rigid()
True
The dimension in which its rigidity
is supposed to be investigated can be specified via the dim keyword.
G_TP.is_rigid(dim=1)
True
For dimensions greater than 2, no combinatorial rigidity criterion is known. To still get a result, a randomized algorithm can be called using the command:
G_TP.is_rigid(dim=3, algorithm="randomized")
False
In fact, we can compute the maximal dimension, in which a graph is rigid using the method max_rigid_dimension().
This method uses a randomized algorithm to determine rigidity in an arbitrary dimension, so it throws a RandomizedAlgorithmWarning. To silence it, we can set Graph.silence_rand_alg_warns=True.
Graph.silence_rand_alg_warns = True
G_TP.max_rigid_dimension()
2
Furthermore, we can compute the rigid components of a graph using rigid_components(),
which is returned as a partition of vertices.
G = graphs.DoubleBanana()
G.rigid_components(dim=3, algorithm="randomized")
[[0, 1, 2, 3, 4], [0, 1, 5, 6, 7]]
We can also investigate the (generic) (global)
rigidity of a graph using the method is_globally_rigid():
G_TP.is_globally_rigid()
False
and
G4 = graphs.ThreePrismPlusEdge()
G4.plot()
G4.is_globally_rigid()
True
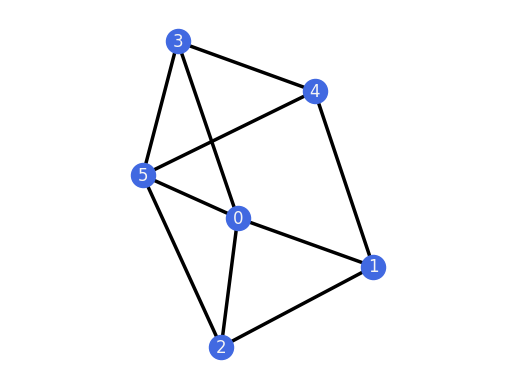
To distinguish graphs from frameworks in the plot() method, the vertices are colored differently by default.
The graph G4 obtained by adding an edge to the 3-prism is globally rigid because it is 3-connected and
redundantly rigid. The redundant rigidity of a graph can be checked via is_redundantly_rigid().
G4.is_redundantly_rigid()
True
We can also investigate the global rigidity for other dimensions, too.
G_TP.is_globally_rigid(dim=1)
True
Finally, it may be useful to check whether a graph can be constructed via an extension sequence,
which can be done using the method extension_sequence().
for H in G_TP.extension_sequence(return_type="graphs"):
H.plot(canvas_height=2)
We can obtain all non-isomorphic \(k\)-extensions of a graph using the method all_k_extensions().
For the 3-prism we ensure that all of the 0-extensions are rigid:
for H in G_TP.all_k_extensions(0, only_non_isomorphic=True):
H.plot()
assert(H.is_rigid())
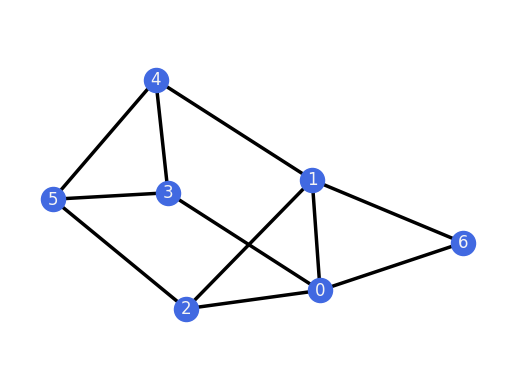
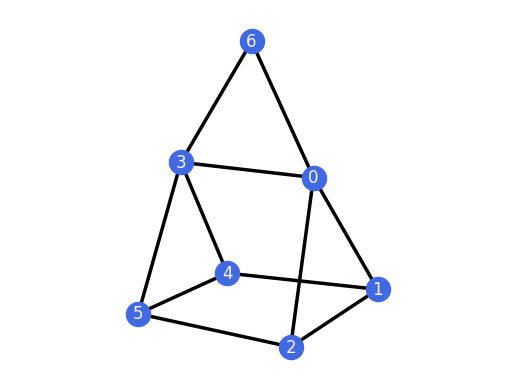
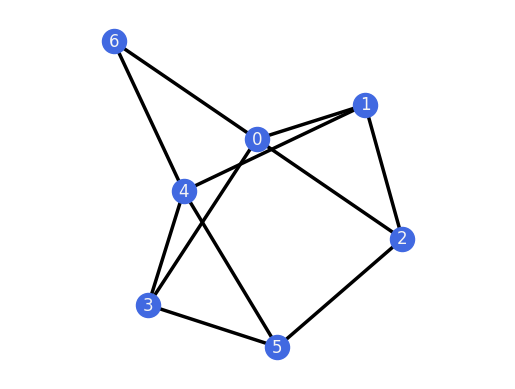
And we can do the same for the 1-extensions of the 3-prism.
for H in graphs.ThreePrism().all_k_extensions(1, only_non_isomorphic=True):
H.plot()
assert(H.is_rigid())
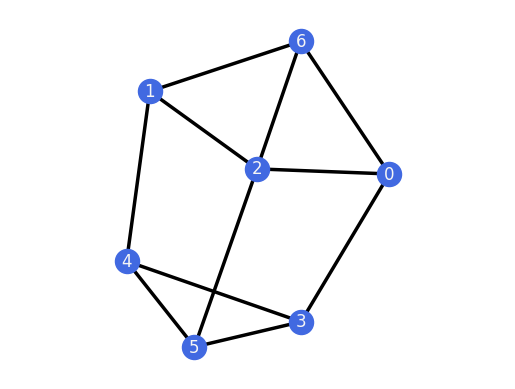
Sparsity¶
The (k,l)-sparsity of a graph can be checked using the method is_kl_sparse().
G = graphs.CompleteBipartite(3,3)
G.is_kl_sparse(2, 3)
True
Famously, the double banana is (3,6)-sparse, but not rigid.
G = graphs.DoubleBanana()
print(G.is_kl_sparse(3, 6))
print(G.is_rigid(dim=3, algorithm="randomized"))
True
False
Similarly, it can be checked whether a graph is (k,l)-tight.
TP = graphs.ThreePrism()
TP.is_kl_tight(2, 3)
True
Matroidal Properties¶
We can use PyRigi to check properties of graphs in the d-dimensional rigidity matroid as well.
To check whether a graph is (in-)dependent in this matroid, we can call
is_Rd_dependent() or is_Rd_independent().
TP.is_Rd_independent()
True
It can be checked whether a graph is a circuit in the d-dimensional rigidity matroid by calling is_Rd_circuit().
TP.is_Rd_circuit()
False
Finally, it can be determined whether the graph is \(R_d\)-closed by calling is_Rd_closed().
TP.is_Rd_closed(dim=1)
False